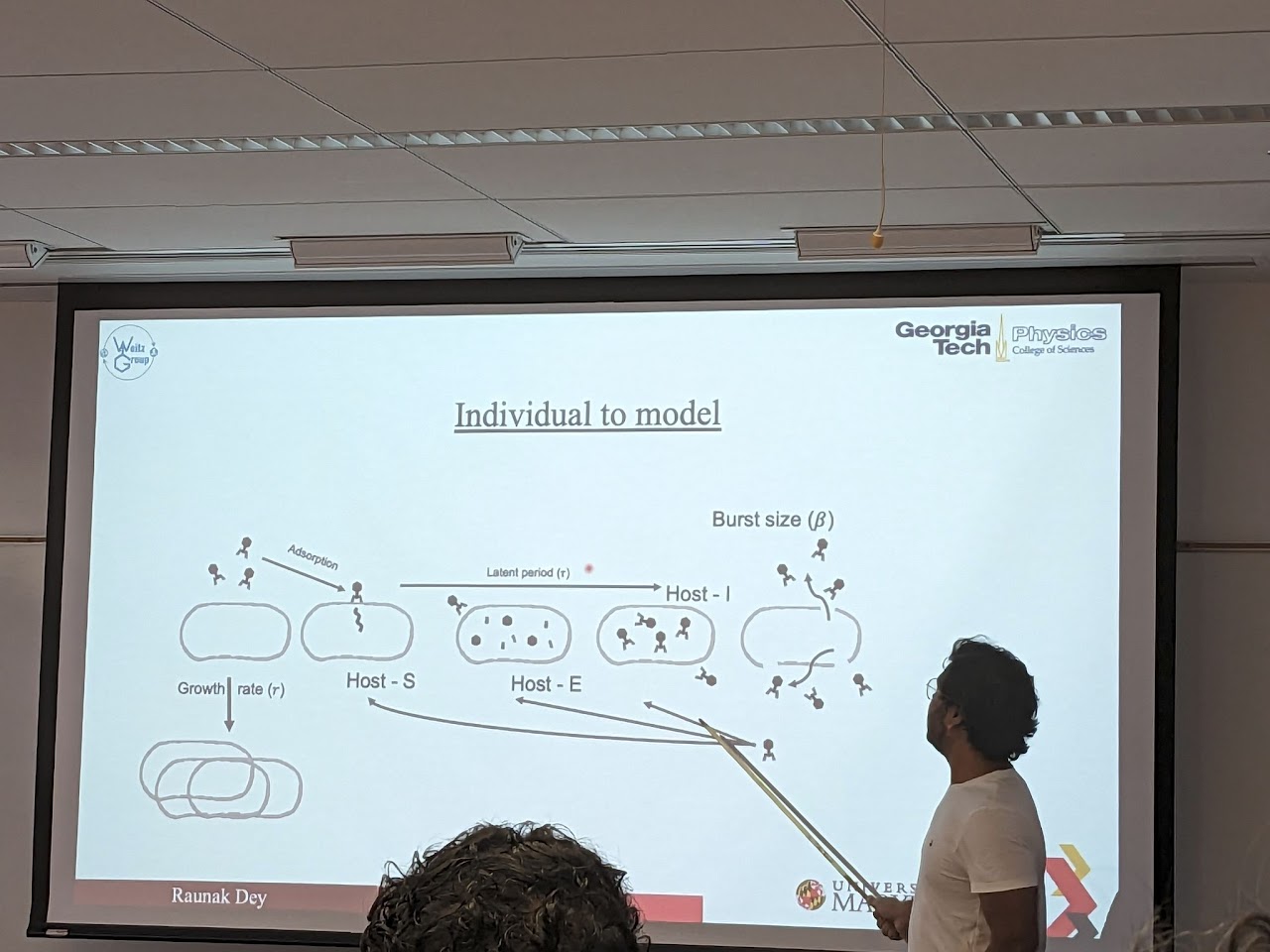
Phage Therapy: Some viruses (phages) can infect and kill bacterial cells, and are hence used for treating resistant infections. We design pharmacological models of phage therapy using phage cocktails and antibiotics. Aided by HFIM data, we use inverse models to inform such infection treatment models and ask the question: is combination therapy better than single therapy? what is the optimal timing and dosage of such treatments?
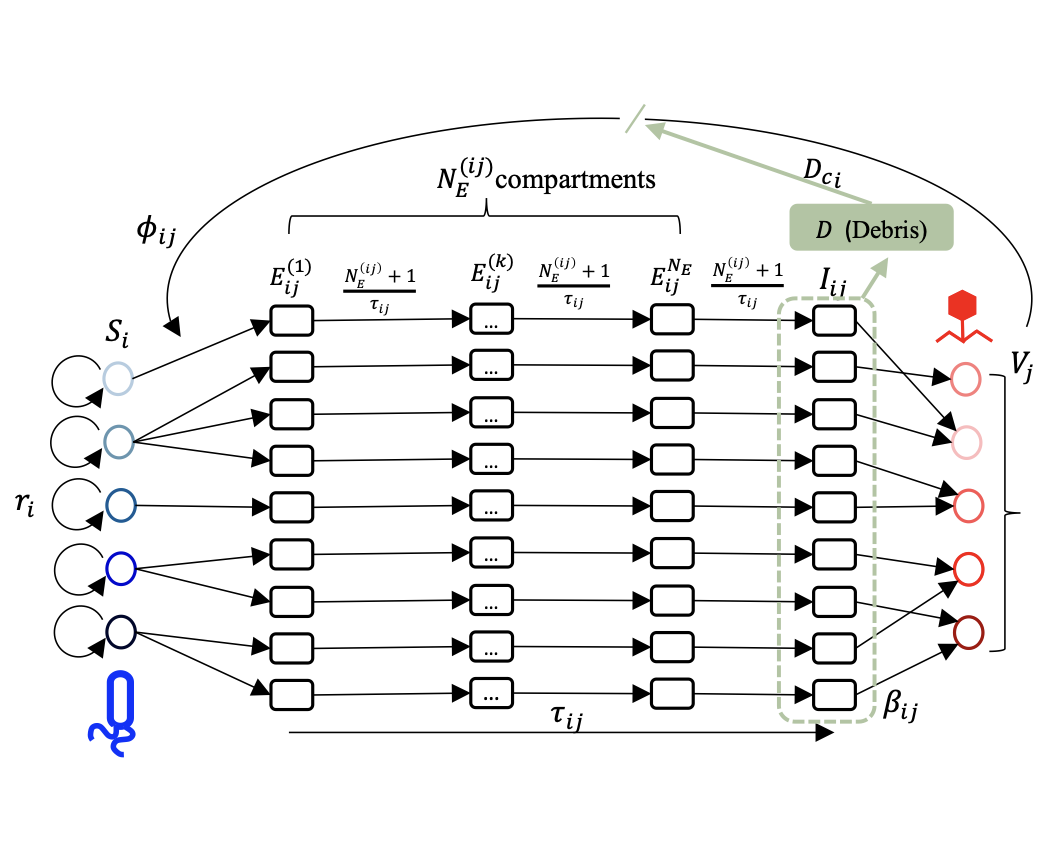
Network of community infections: In natural ecosystems, multiple species of phages coexist with their bacterial hosts. From observed population dynamics data, we address the question: which phage infects which microbes? Which microbial groups cooperate vs compete? How to infer underlying traits from the observed population dynamics? How different are pairwise interactions from community interactions? How are the populations stabilized in a natural ecosystem? We use ODEs and interaction models to parametrize these systems and constrained optimization techniques to solve inverse problems in these complex systems.
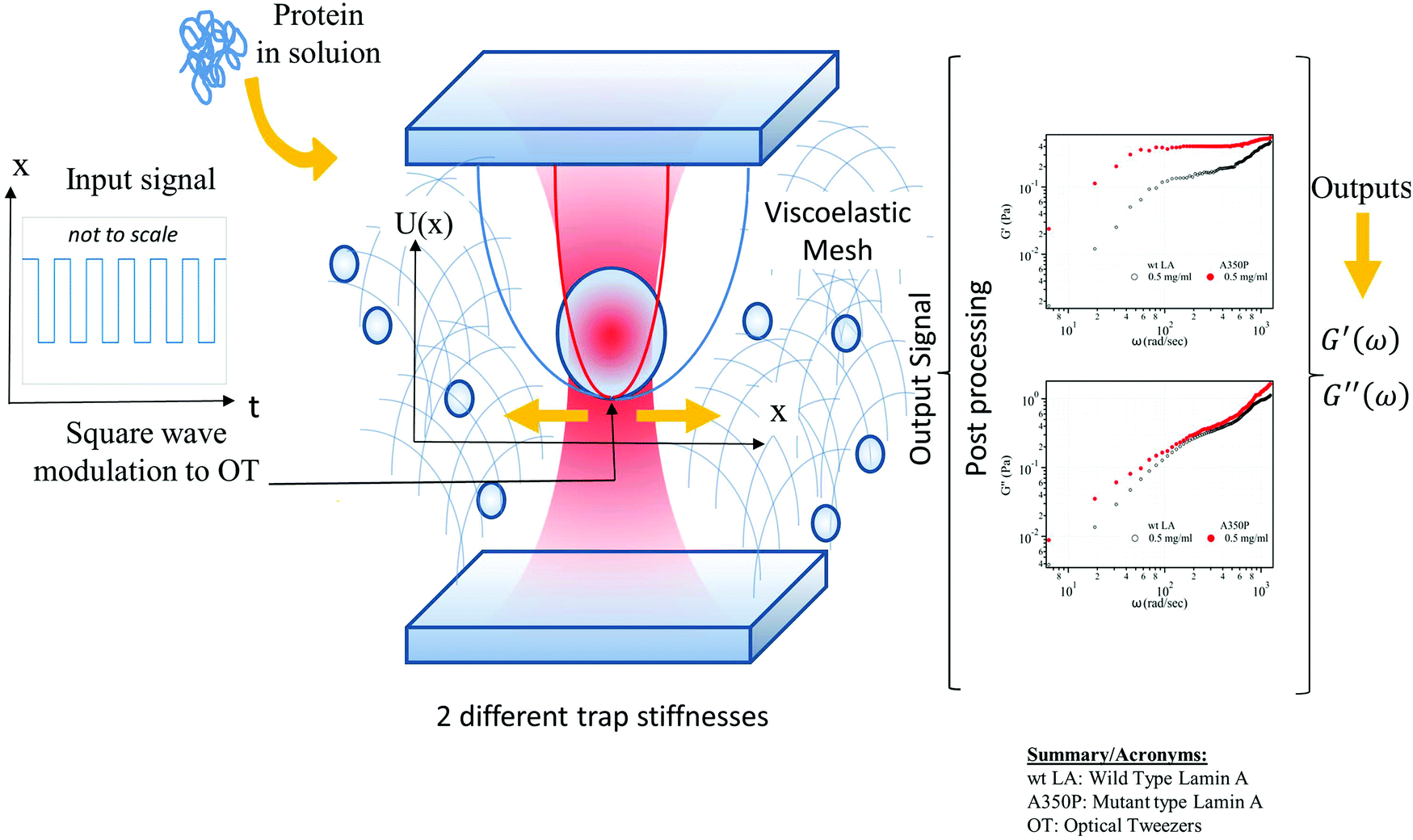
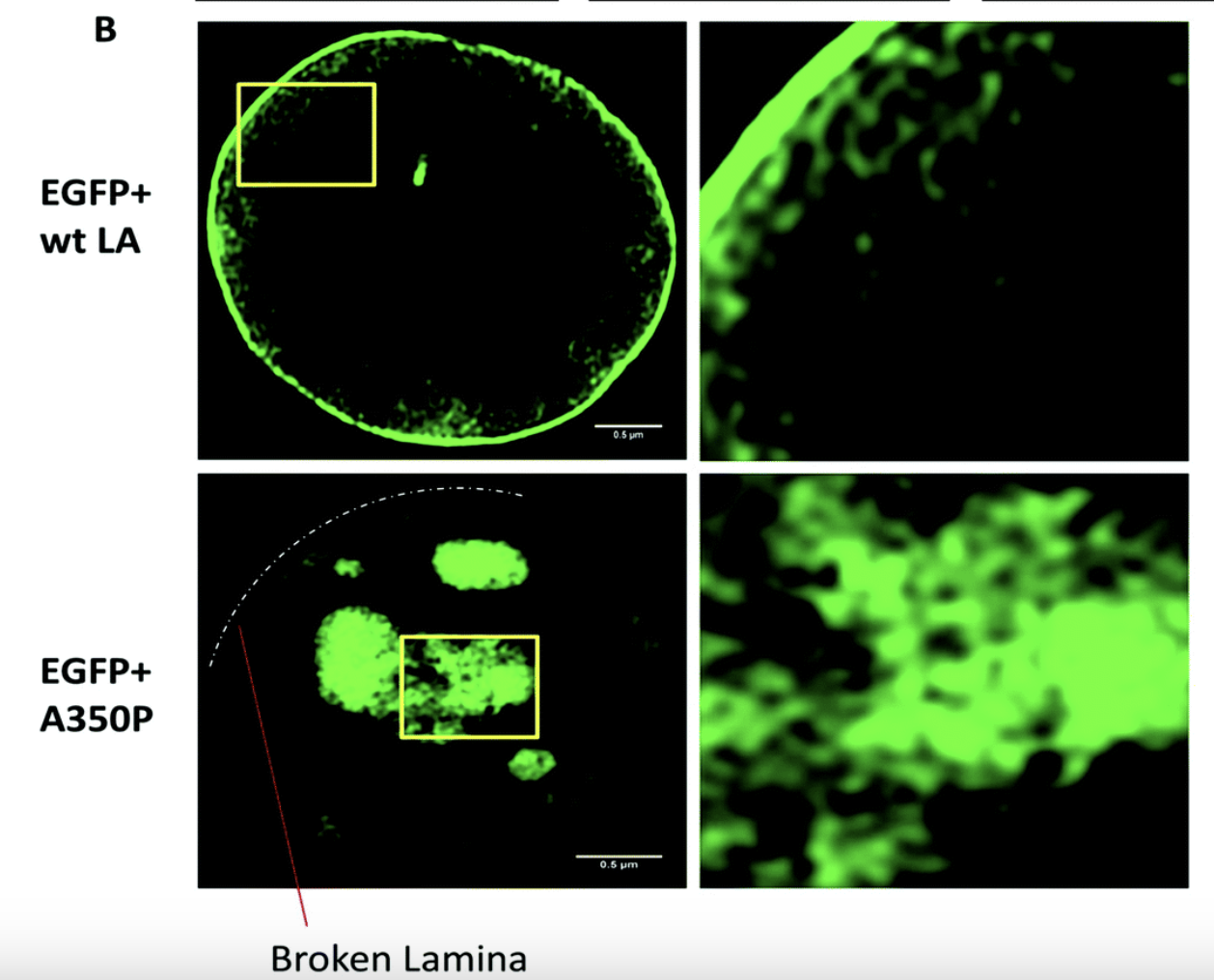
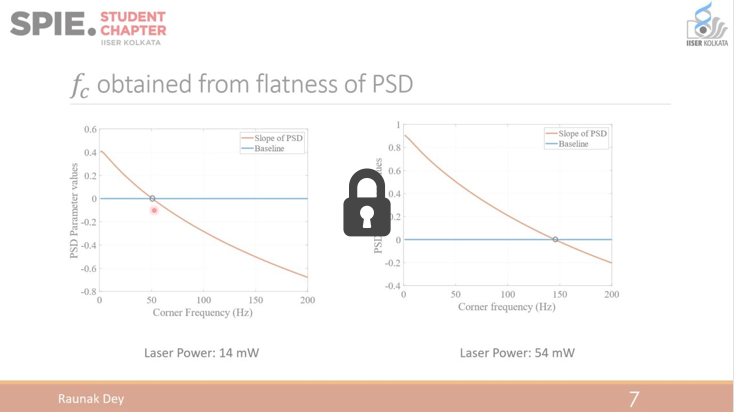
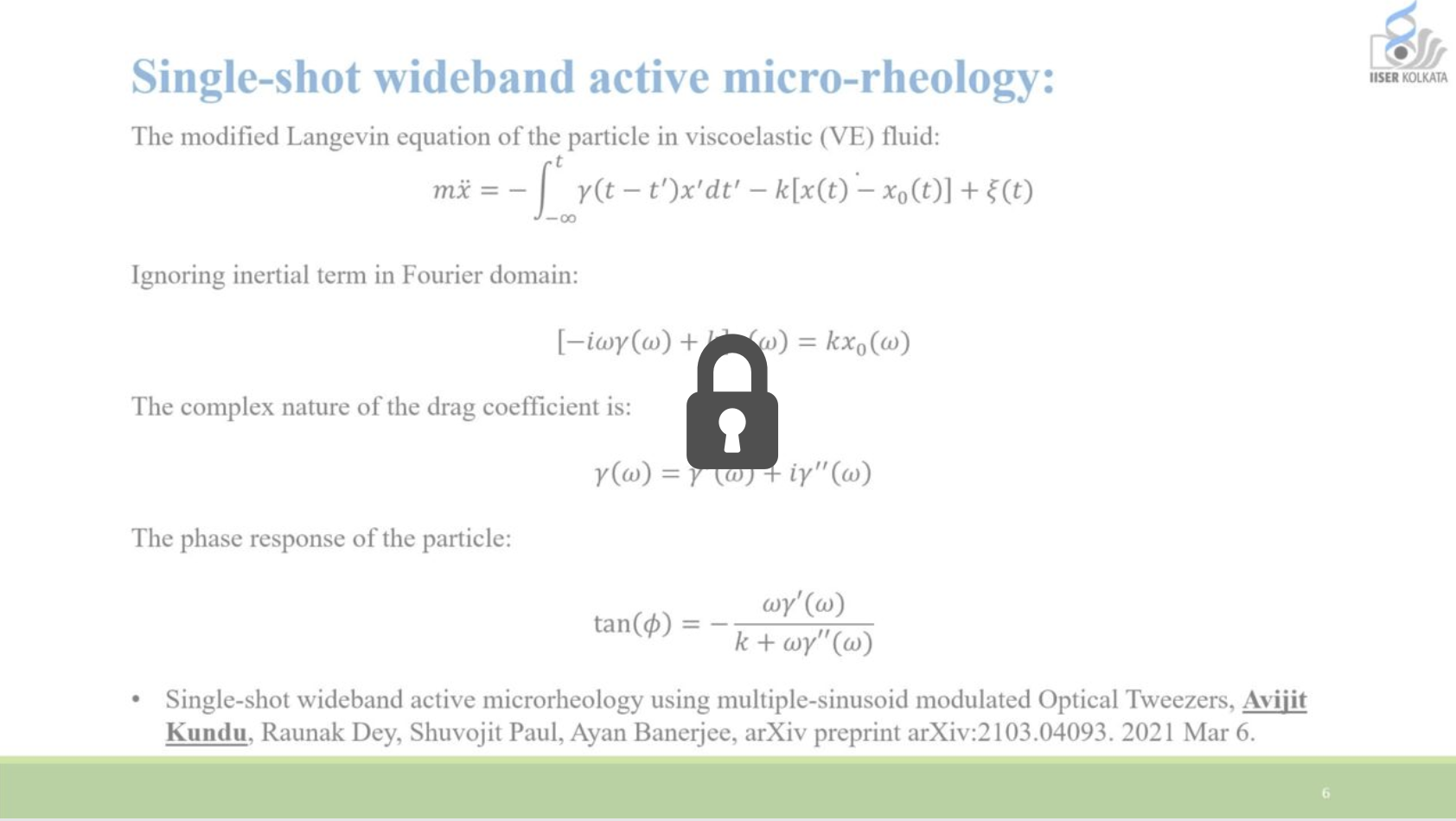
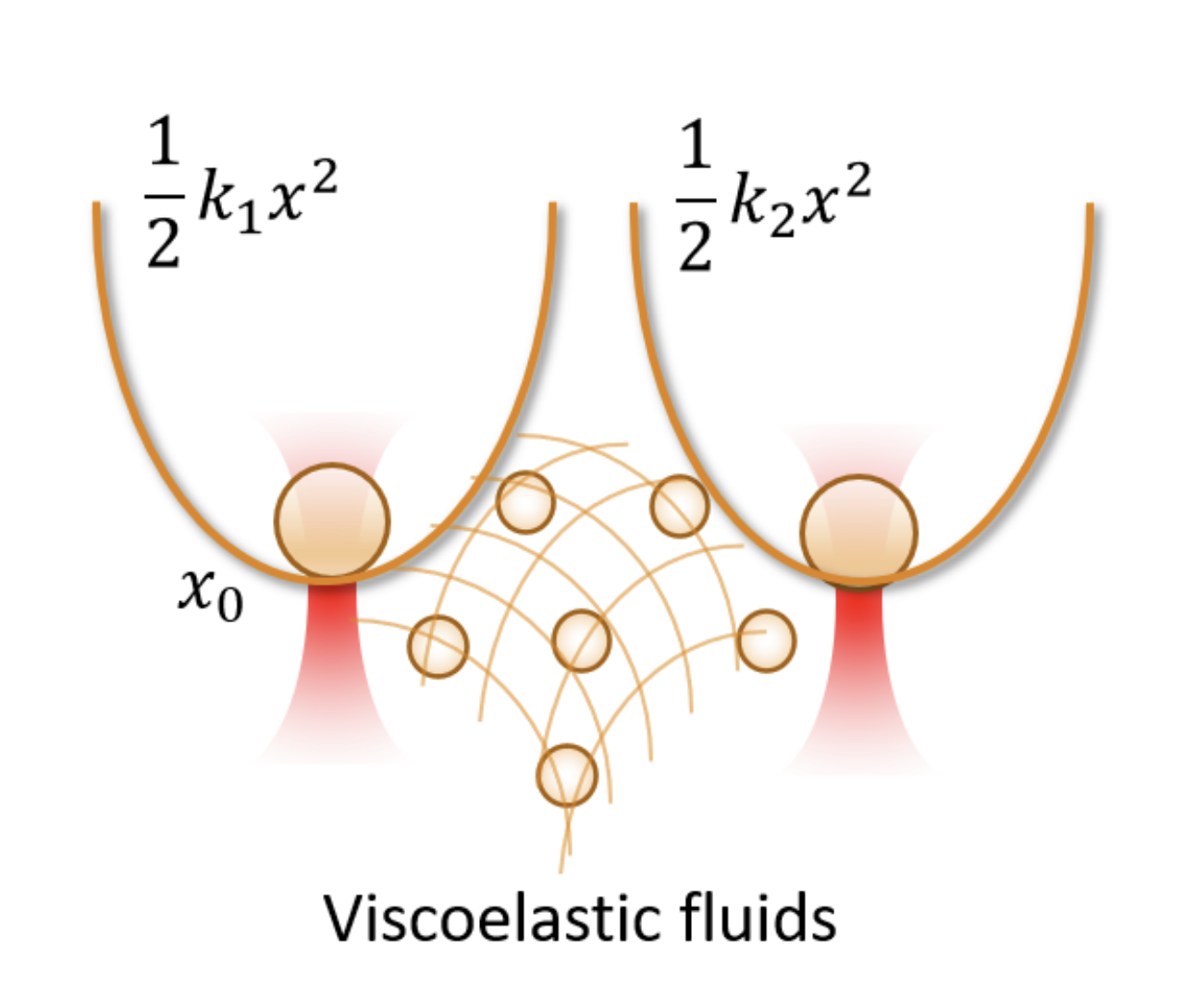
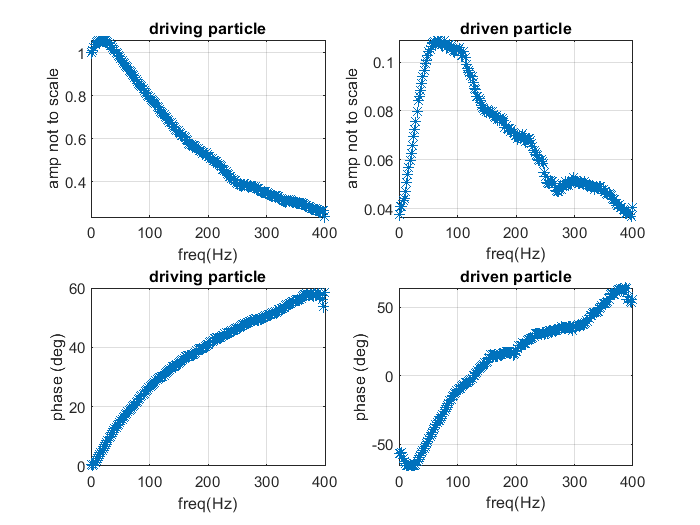
Optimal sensing via microrheology: In experiments guided with Optical Tweezers, a submicroscopic probe is embedded in complex non-Newtonian fluid. From the observed motion of the microscopic probe, the rheological properties of the fluid can be determined. We design(ed) optimal control algorithms to optimize the signal-to-noise ratio in wideband response measurements, such that they remain in a linear-response regime. Our constrained optimization algorithm outperforms standard techniques in terms of speed and accuracy, which is essential for time-sensitive experiments with biological media. We have used these to study how viscoelastic properties of mutated Lamin differ from the wild type, a mutation linked to cardiomyopathy.
Sequential Bayesian localization of acoustic emitter via mobile drone: Drones are widely used in search and rescue. Through numerical simulations, we devised a sequential Bayesian framework, for optimally moving a receiver drone to localize an acoustic emitter. The drone uses phase difference of arrival from the source at different time intervals, and each step identifies a posterior for possible location; then it computes where to optimally move. (Hint: moving directly towards the source is not optimal.)
LLM-based time series forecasting: LLMs being trained on large datasets have reliable autoregressive next-word prediction capabilities. Tokenizers are being developed where numbers are mapped to tokens and solving the next token estimation problems with pre-trained LLMs solves the next number prediction problem for time series. For different LLMs and tokenizations, we investigate how zero-shot or few-shot learning is effective for chaotic and stochastic systems (Hint: it is not!).
Gen-AI for time series extrapolation: Gen-AI models such as VAE, GAN, and diffusion models are excellent in approximating the underlying probability distributions from samples. For physics-derived dynamical systems, we explore Gen-AI models for predictive tasks for undersampled data and sparse time series.
Real random numbers from Brownian trajectories:
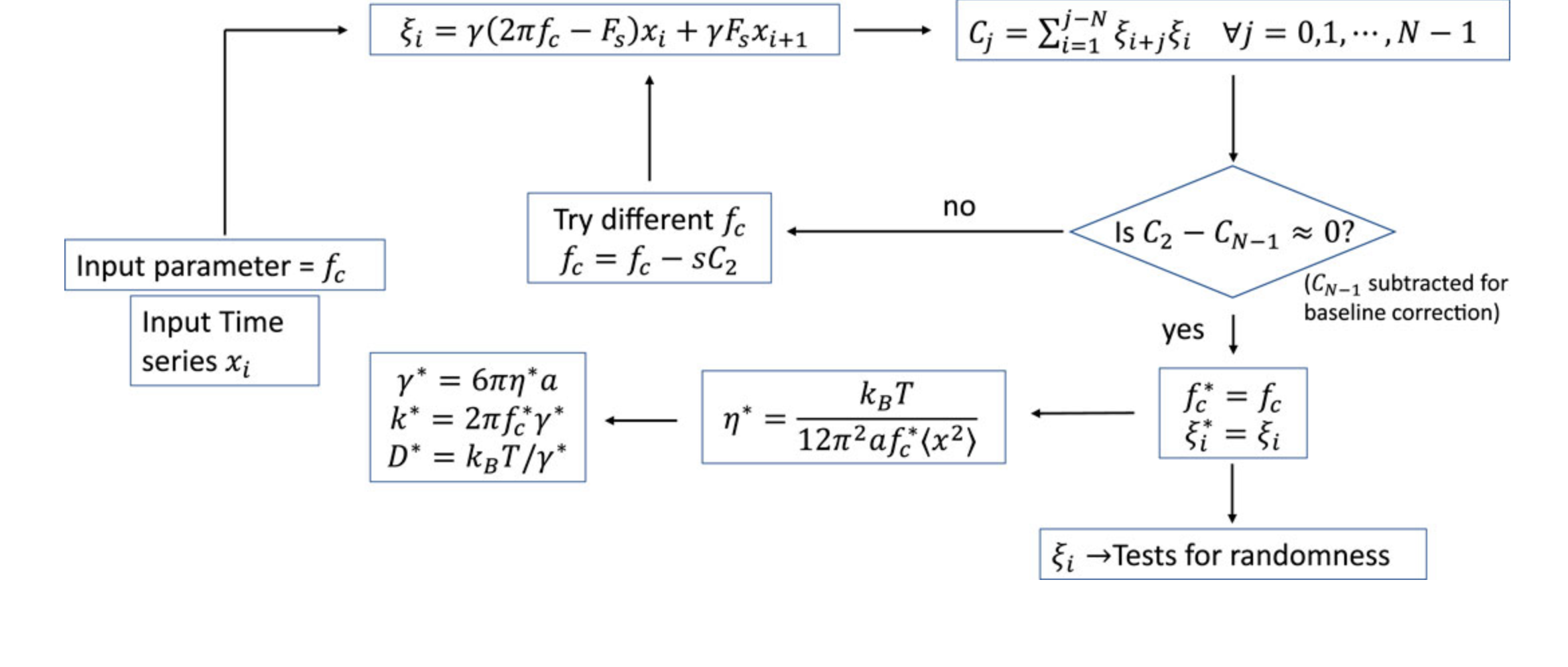
Algorithms for random number generation produce pseudo-random numbers. Through experimental trajectories of Brownian probes captured via our optical tweezers setup, we developed random number extraction algorithms, based on NIST tests for randomness. We found that the entropy of randomness improves asymptotically as the sampling rate increases.
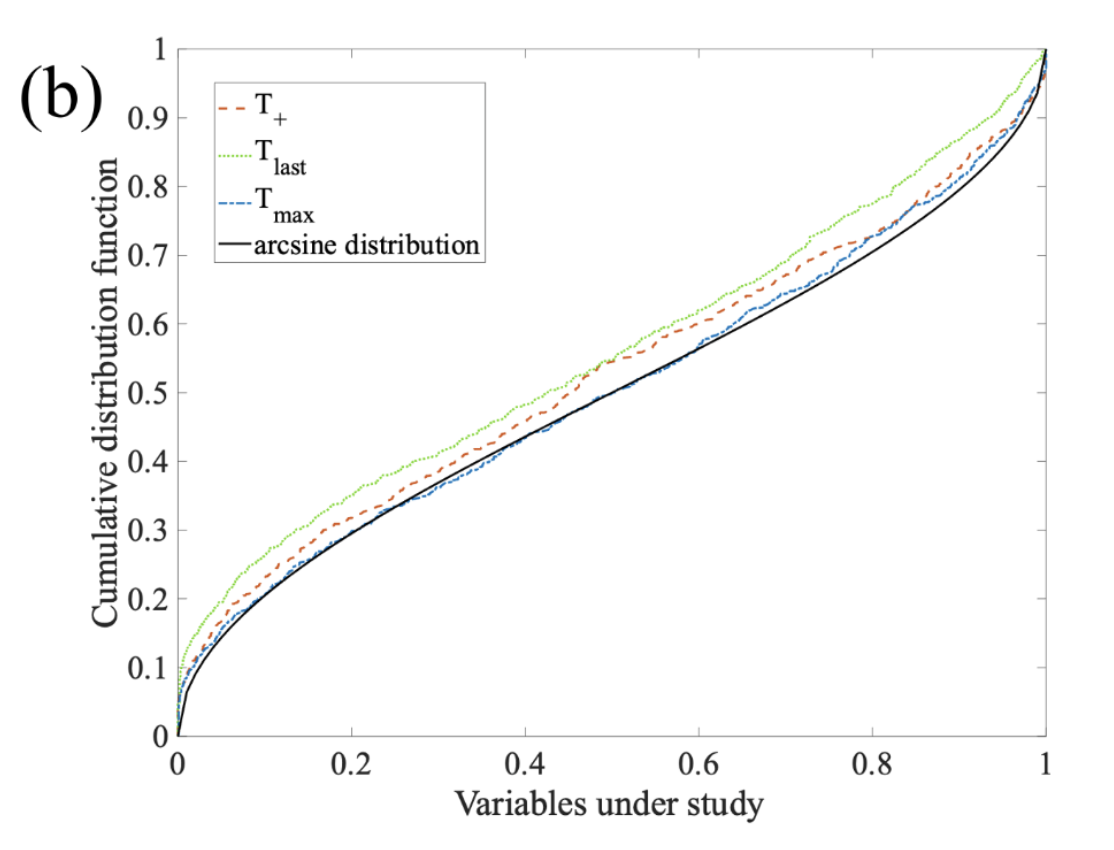
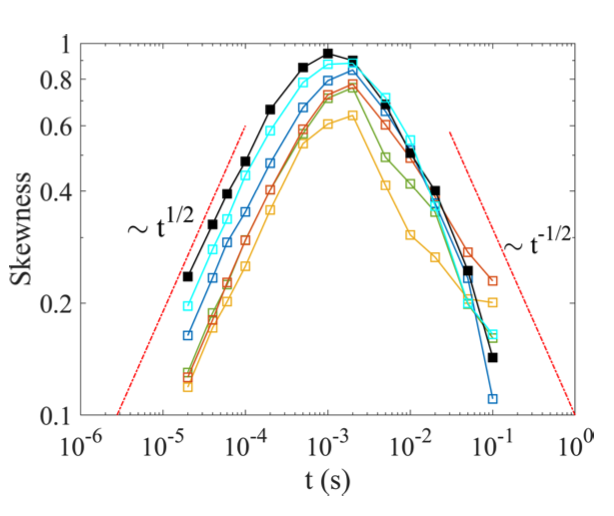
Non-equilibrium statistical mechanics: We study energy dissipation and thermodynamic currents driving stochastic probes in fluids. With experimentally recorded Brownian trajectories, we discovered how these currents converge to an arcsine distribution, and how they have directional preference depending on the timescale of observation.
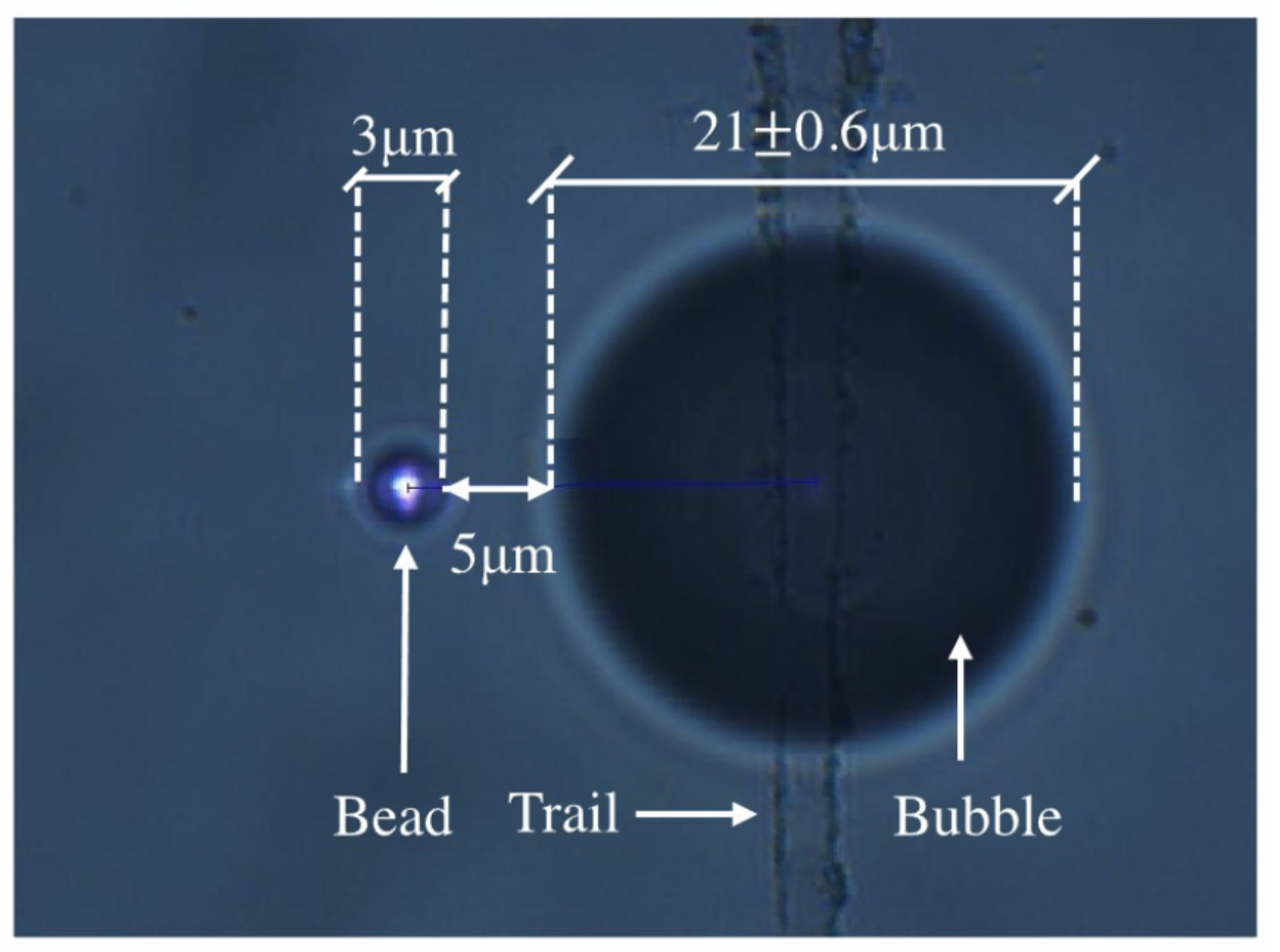
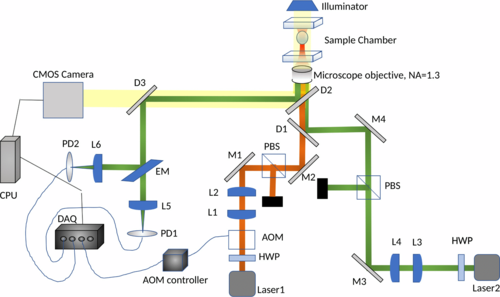
Optical Tweezers: A collimated beam of laser when focused by a high numerical aperture lens can capture microscopic probes, such that they can be moved along with the laser beam. At the Light-Matter Lab (IISER Kolkata) , I developed custom-built optical tweezers and imaging systems for the simultaneous capture of multiple sub-microscopic particles; along with modulation and tracking with nanometer resolution.
Quantum Spectroscopy: Entangled photons interacting with matter have their properties altered due to light-matter interactions. At the Silva Lab (Georgia Tech), I worked on building time-synchronized dual single-photon detection systems, for quantum metrology with these entangled photon pairs.
Microscopy and imaging: At the Li Lab in the University of British Columbia, I build a custom microscope with two high numerical aperture lenses and 3D motor controls enabled by gaming joysticks.
GMRT (Giant Meterwave Radio Telescope) in India, employs up to 33 radio telescope dishes to image pulsars (rapidly rotating neutron stars that emit beams of electromagnetic radiation). Due to RFID, these frames (lasting only a few seconds have low SNR). Using multi-detector observations and stacking, we developed a pipeline to filter RDIF corrupt data for radio frequency observation.